Recombinant Rat CLOCK Protein
| Cat.No. : | CLOCK-1459R |
| Product Overview : | Recombinant Rat CLOCK full length or partial length protein was expressed. |
- Specification
- Gene Information
- Related Products
- Citation
- Download
| Species : | Rat |
| Source : | Mammalian Cells |
| Tag : | His |
| Form : | Liquid or lyophilized powder |
| Endotoxin : | < 1.0 EU per μg of the protein as determined by the LAL method. |
| Purity : | >80% |
| Notes : | This item requires custom production and lead time is between 5-9 weeks. We can custom produce according to your specifications. |
| Storage : | Store it at +4 ºC for short term. For long term storage, store it at -20 ºC~-80 ºC. |
| Storage Buffer : | PBS buffer |
| Gene Name | Clock clock homolog (mouse) [ Rattus norvegicus ] |
| Official Symbol | CLOCK |
| Gene ID | 60447 |
| mRNA Refseq | NM_021856.1 |
| Protein Refseq | NP_068628.1 |
| MIM | |
| UniProt ID | Q9WVS9 |
Diurnal oscillations of endogenous H 2 O 2 sustained by p66 Shc regulate circadian clocks
Journal: Nature cell biology PubMed ID: 31768048 Data: 2020/4/27
Authors: Jian-Fei Pei, Xun-Kai Li, De-Pei Liu
Article Snippet:One microgram of recombinant CLOCK protein (Creative BioMart) was incubated with 1 mM hydrogen peroxide for 10 min at 37°C, at which point dimedone was added to a final concentration of 5 mM and the samples were incubated for an additional 1 h. After the dimedone treatment, 10 mM DTT was added, and the samples were incubated for an additional 30 min at 37°C.. Subsequently, samples were incubated with 20 mM indole-3-acetic acid (IAA) for an additional 30 min, after which trypsin digestion was performed overnight.Subsequently, samples were incubated with 20 mM indole-3-acetic acid (IAA) for an additional 30 min, after which trypsin digestion was performed overnight.
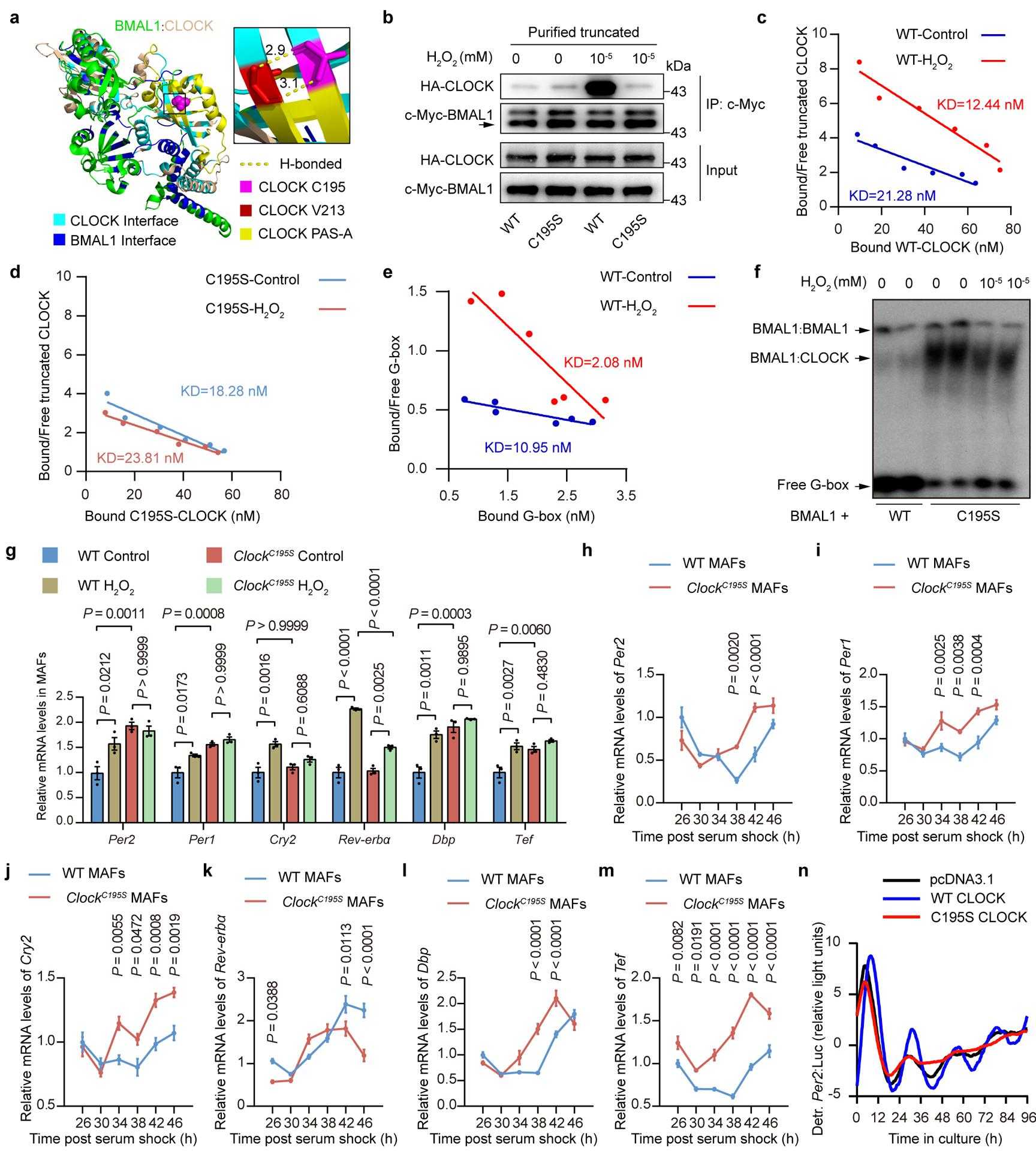
a, Environment of the C195 thiol in the
Not For Human Consumption!
Inquiry
- Reviews (0)
- Q&As (0)
Ask a Question for All CLOCK Products
Required fields are marked with *
My Review for All CLOCK Products
Required fields are marked with *



