Alzheimer's Disease
Related Symbol Search List
- COLQ
- ADAM10
- ADAM17
- ADAM9
- RAGE
- APH1A
- APP
- BACE1
- BECN1
- Caspase-3
- CASP6
- CASP8
- CASP9
- GABRA1
- GNRH1
- GRIN2C
- GRM5
- GSK3B
- HSP90AA1
- MAPT
- MTOR
- NFE2L2
- NGF
- PDE4A
- PDE5A
- PLD2
- PPARG
- PPARGC1A
- PSEN1
- PSEN2
- PSENEN
- PTGS1
- Ppp2r4
- SIRT2
Immunology Background
About Alzheimer's Disease and Related Molecules
Alzheimer's disease (AD) is a neurodegenerative disorder that primarily affects older adults, causing progressive cognitive decline and memory loss. It is the most common cause of dementia and is characterized by the accumulation of abnormal proteins in the brain, which leads to the loss of neurons and synaptic connections, and causes the death of brain cells.
Key molecules associated with Alzheimer's disease are beta-amyloid, tau proteins, and others. These include inflammatory proteins such as cytokines and chemokines, oxidative stress-related molecules, and neurotransmitters such as acetylcholine, which are important for memory and cognition.
Understanding the role of these molecules in Alzheimer's disease is crucial for developing effective treatments. Current research focuses on developing drugs that can target beta-amyloid and tau to prevent their accumulation or promote their clearance. Other strategies include reducing inflammation, improving brain cell function, and finding ways to protect and repair damaged neurons.
Overall, Alzheimer's disease is a complex disorder involving multiple molecules and pathways. While much progress has been made in understanding its causes, there is still much to learn to find a cure or effective treatment for this devastating disease.
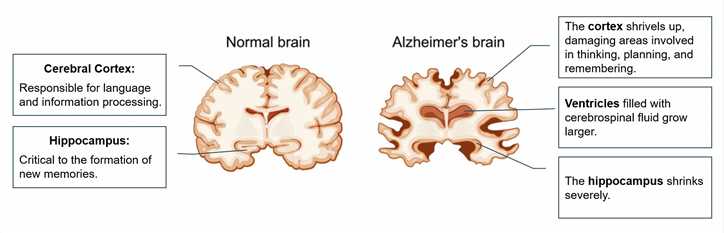
Biological Functions of Alzheimer's Disease-Related Molecules
Alzheimer's disease is a neurodegenerative disorder characterized by the progressive loss of brain function, particularly in areas involved in memory and cognitive abilities. Several molecules have been identified as being involved in the development and progression of Alzheimer's disease, and understanding their biological functions is crucial for developing potential therapeutic interventions. Here are some of the biological functions of Alzheimer's disease-related molecules:
Amyloid-beta (Aβ): Aβ peptides are derived from the amyloid precursor protein (APP) and are known to accumulate in the brains of individuals with Alzheimer's disease. The major biological function of Aβ is believed to be its involvement in the formation of senile plaques, which are one of the hallmarks of Alzheimer's disease. Aβ accumulation disrupts normal brain function, leading to neuronal dysfunction and ultimately cell death.
Tau Protein: Tau is a microtubule-associated protein that plays a crucial role in stabilizing microtubules in neurons. In Alzheimer's disease, abnormal tau protein aggregates to form neurofibrillary tangles, another characteristic feature of the disease. The accumulation of these tangles disrupts the normal functioning of neurons and impairs their ability to transport essential molecules within the cell.
Apolipoprotein E (APOE): APOE is a protein involved in lipid metabolism, and it plays a vital role in transporting cholesterol in the brain. There are three common forms of the APOE gene, with the APOE4 allele being the strongest genetic risk factor for late-onset Alzheimer's disease. The exact biological function of APOE in Alzheimer's disease is still not fully understood, but it is believed that APOE4 may influence Aβ aggregation and clearance, synaptic plasticity, and neuroinflammation.
Presenilin 1 and Presenilin 2: Mutations in the presenilin 1 and presenilin 2 genes are associated with early-onset familial Alzheimer's disease. Presenilin 1 and presenilin 2 proteins are transmembrane proteins that are localized in the endoplasmic reticulum and Golgi apparatus. They are responsible for the catalytic activity of the gamma-secretase complex, which cleaves the transmembrane domain of APP to release amyloid-beta peptides. Mutations in the presenilin genes can lead to increased production and accumulation of toxic forms of amyloid-beta peptides, particularly the amyloid-beta 42 peptide. This accumulation leads to the formation of beta-amyloid plaques, which are a characteristic feature of Alzheimer's disease.
Understanding the normal biological functions of these molecules provides insight into their roles in maintaining neuronal health and function. However, in the context of AD, dysregulation and pathological changes in these molecules contribute to the characteristic features of the disease, such as beta-amyloid plaques, tau tangles, and cholinergic dysfunction. Targeting these molecules and their associated pathways is an important approach to developing potential therapies for AD.
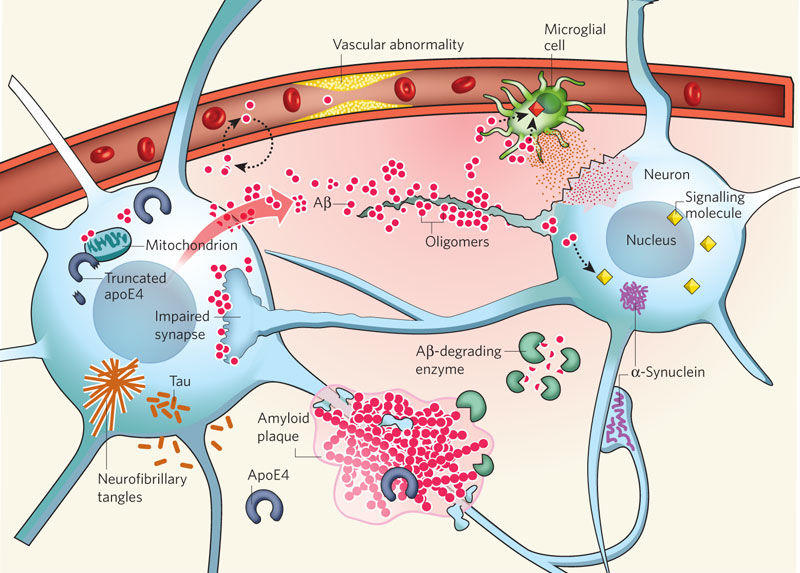 Fig.2 Alzheimer's disease: pathophysiology (Some key players in the pathogenesis of AD). (Mucke L, 2009)
Fig.2 Alzheimer's disease: pathophysiology (Some key players in the pathogenesis of AD). (Mucke L, 2009)
Implications for Studying Alzheimer's Disease-related Molecular
Alzheimer's disease (AD) is a progressive neurodegenerative disorder that affects millions of people worldwide. As scientists continue to unravel the molecular mechanisms underlying this devastating disease, significant advancements have been made in AD-related molecular research that have paved the way for potential therapeutic interventions. The significance and application areas of such research are substantial and offer hope for the development of effective treatments for AD patients.
Understanding Disease Progression: Molecular research allows scientists to explore the intricate cellular and molecular processes associated with AD. By deciphering the underlying mechanisms, researchers can better understand the progression of the disease and identify potential targets for intervention.
Biomarkers: Identification and characterization of AD-related molecular biomarkers aid in the diagnosis and monitoring of the disease. Biomarkers can be used to detect AD in its early stages, track disease progression, and evaluate the response to treatments. Molecular biomarkers, including beta-amyloid and tau proteins, can be measured in cerebrospinal fluid, blood, or through neuroimaging techniques, enabling improved diagnostic accuracy and personalized patient management.
Drug Discovery and Development: Molecular research provides insights into potential therapeutic targets for drug development. By understanding the molecular pathways involved in AD pathology, scientists can design and test novel drugs that can modulate these pathways and potentially slow down or halt disease progression. This research has led to the development of drugs targeting Aβ, tau, neuroinflammation, and other AD-related molecular processes.
Therapeutic Targets: AD-related molecules serve as potential therapeutic targets for the development of treatments. Understanding the roles of these molecules, such as beta-amyloid and tau, in AD pathogenesis allows researchers to design interventions aimed at preventing their accumulation, promoting their clearance, or mitigating their toxic effects. Targeting these molecules and associated pathways may help in developing disease-modifying therapies to slow down or halt disease progression.
Available Resources for Alzheimer's Disease
Creative BioMart offers a broad range of products and services to support research into Alzheimer's disease and related molecules. Our offerings include recombinant proteins, cell and tissue lysates, protein pre-coupled beads, and others. We also offer custom services to meet specific research needs. Additionally, we provide resource support, including pathways, protein functions, interacting proteins, and relevant articles, to enhance understanding and study of these molecules. Explore the Alzheimer's disease-related molecules below for more comprehensive resources.
We are dedicated to providing you with high-quality research tools and services to help you achieve successful scientific outcomes. If you have any further questions or require custom services, please feel free to contact us at any time.
References:
- Fan L, Mao C, Hu X, et al. New Insights Into the Pathogenesis of Alzheimer's Disease. Front Neurol. 2020;10:1312. Published 2020 Jan 10.
- Chen XQ, Mobley WC. Alzheimer Disease Pathogenesis: Insights From Molecular and Cellular Biology Studies of Oligomeric Aβ and Tau Species. Front Neurosci. 2019;13:659.
- Mucke L. Neuroscience: Alzheimer's disease. Nature. 2009;461(7266):895-897.

