Hematopoietic Stem Cell Transcription Factors and Regulators
Related Symbol Search List
- AHR
- CDX4
- DNMT3A
- DNMT3B
- EGR1
- GATA2
- GATA3
- HHEX
- HMGB1
- HMGB3
- IKZF2
- JUN
- LMO2
- LMO4
- MAF
- MAFB
- MEF2C
- MYB
- MYC
- NFATC2
- NFE2L2
- PITX2
- PRDM16
- PROX1
- RUNX1
- SALL4
- SETD2
- smad7
- SPI1
- STAT3
- STAT4
- STAT6
- TAL1
- p53
- TSC22D1
Immunology Background
Overview of Hematopoietic Stem Cell Transcription Factors and Regulators
Hematopoietic stem cells (HSCs) are multipotent stem cells, which can give rise to all types of blood cells including myeloid (monocytes, macrophages, neutrophils, basophils) and blood cell types. macrophages, neutrophils, basophils, eosinophils, erythrocytes, megakaryocytes/platelets, and dendritic cells) and lymphoid lineages (T cells, B cells, NK cells). The regulatory mechanism of HSCs involves the synergistic action of multiple transcription factors and regulators. These transcription and regulatory factors are involved in the regulation of HSC self-renewal, proliferation, and differentiation, and are essential for maintaining the function and homeostasis of the hematopoietic system. In addition, the study of these factors offers the possibility of developing clinically applicable ex vivo expansion methods that are expected to generate sufficient numbers of HSCs for the treatment of various diseases. Some important HSC transcription factors, such as GATA-2, RUNX1, SCL/TAL1, FLI1, and MYB, as well as some transcriptional regulators, such as HOXB4, NOTCH, Wnt, and BMP, affect the normal development and function of the hematopoietic system by regulating the processes of HSC proliferation, differentiation, and self-renewal. Studying the functions and interactions of HSC transcription factors and regulators is of great significance in gaining insight into the regulatory mechanisms of hematopoietic processes, stem cell therapy, and the occurrence and treatment of diseases related to the hematopoietic system.
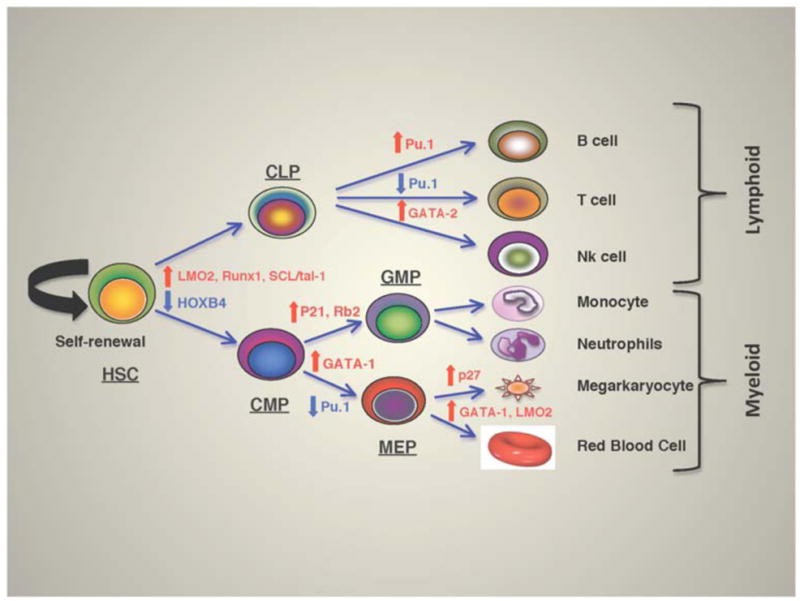 Fig.1 Transcriptional regulation of hematopoiesis. Crosstalks of various transcriptional factors regulate the self-renewal HSCs or commit to either lymphoid or myeloid lineages depending on the expression levels. (Aggarwal R, et al., 2012)
Fig.1 Transcriptional regulation of hematopoiesis. Crosstalks of various transcriptional factors regulate the self-renewal HSCs or commit to either lymphoid or myeloid lineages depending on the expression levels. (Aggarwal R, et al., 2012)
Functions of Hematopoietic Stem Cell Transcription Factors and Regulators
Hematopoietic stem cell transcription factors and regulators play multiple specific roles in regulating the function and fate determination of hematopoietic stem cells (HSCs).
- Self-renewal regulation
The study found that the transcription factor GATA-2 plays an important role in regulating the self-renewal process of HSCs. Using a conditional GATA-2 knockout mouse model, the researchers found that deletion of GATA-2 leads to a reduction in the number of HSCs and impaired function. The study also showed that GATA-2 promotes self-renewal of HSCs by regulating their proliferation and maintaining stemness characteristics (Tsai et al., 1994).
- Regulation of differentiation
It has been found that deletion of RUNX1 leads to abnormal differentiation and loss of hematopoietic function in HSCs. By using a RUNX1 knockout mouse model, the researchers found that RUNX1 regulates the expression of multiple genes, including those related to hematopoiesis and differentiation. These results suggest that RUNX1 is a key regulator of differentiation of HSCs (Wang et al., 1996).
- Maintenance of stem cell properties
HOXB4 is an important transcriptional regulator that is essential for the maintenance of stem cell properties of HSCs. Studies have shown that HOXB4 maintains the stemness characteristics of HSCs by regulating the expression of several genes, including stem cell-related genes. By studying HOXB4 overexpressing mice, researchers have found that HOXB4 increases the number and self-renewal capacity of HSCs while inhibiting their differentiation into specific blood cell lineages (Antonchuk et al., 2002).
- Fate-determined regulation
The NOTCH signaling pathway plays an important role in the fate decision of HSCs. Activation of the NOTCH signaling pathway was found to maintain the stemness characteristics of HSCs and inhibit their differentiation to specific blood cell families. Through the use of NOTCH signaling pathway modulators and knockout mouse models, researchers have found that the NOTCH signaling pathway influences the proliferation and differentiation process of HSCs by modulating the expression of multiple transcription factors and regulators (Delaney C et al., 2010).
Reference:
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology[J]. Cell, 2008;132(4):631-644.
- Wilkinson A C, Göttgens B. Transcriptional regulation of hematopoietic stem cells[J]. Transcriptional and Translational Regulation of Stem Cells, 2013: 187-212.
- Laurenti E, Göttgens B. From hematopoietic stem cells to complex differentiation landscapes[J]. Nature, 2018, 553(7689): 418-426.
- Tsai F Y, Keller G, Kuo F C, et al. An early hematopoietic defect in mice lacking the transcription factor GATA-2[J]. Nature, 1994, 371(6494): 221-226.
- Wang Q, Stacy T, Binder M, et al. Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis[J]. Proceedings of the National Academy of Sciences, 1996, 93(8): 3444-3449.
- Aggarwal R, Lu J, Pompili VJ, Das H. Hematopoietic stfem cells: transcriptional regulation, ex vivo expansion and clinical application. Curr Mol Med. 2012;12(1):34-49.
- Antonchuk J, Sauvageau G, Humphries R K. HOXB4-induced expansion of adult hematopoietic stem cells ex vivo[J]. Cell, 2002, 109(1): 39-45.
- Rodrigues N P, Boyd A S, Fugazza C, et al. GATA-2 regulates granulocyte-macrophage progenitor cell function[J]. Blood, The Journal of the American Society of Hematology, 2008, 112(13): 4862-4873.
- Growney J D, Shigematsu H, Li Z, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype[J]. Blood, 2005, 106(2): 494-504.
- Delaney C, Heimfeld S, Brashem-Stein C, et al. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution[J].Nature Medicine, 2010, 16(2): 232-236.

