Wnt Intracellular Signaling
Related Symbol Search List
- ROCK2
- ABL1
- ASCL1
- CALM1
- Camk1
- CAMK2A
- CAMK2B
- CaMKII
- CAMK2G
- CSNK1A1
- CSNK1D
- CSNK1E
- CSNK1G1
- CSNK2A1
- CSNK2B
- CTNNB1
- DIXDC1
- FOS
- JUN
- JUND
- MAP3K7
- MAPK10
- MAPK8
- JNK2
- PPP3CA
- PPP3R1
- PPP3R2
- Prkca
- PRKCB
- PRKCD
- PRKCE
- PRKCG
- PRKCI
- GLRX3
- PRKCQ
- PRKCZ
- PRKD2
- PRKD3
- TGFB1I1
- NEDD4L
- UBB
- VANGL2
Immunology Background
Available Resources for Wnt Intracellular Signaling Research
At Creative BioMart you can find a wide range of products related to Wnt intracellular signaling, including recombinant proteins and other key products. In addition, we offer customized services to meet your specific requirements, ensuring you get the product you need.
In addition to our products and services, we offer a wealth of resources for your reference. Our resources cover all aspects of Wnt intracellular signaling, including the involved pathways, protein function, interacting proteins, related articles, research areas, and other relevant topics. These resources will be invaluable to researchers wishing to deepen their understanding of Wnt intracellular signaling and their role in physiological processes.
Our Featured Products
About Wnt Intracellular Signaling
Wnt intracellular signaling refers to the cascade of molecular events that occur inside a cell upon activation of the Wnt signaling pathway. The Wnt pathway is a highly conserved signaling pathway that plays a crucial role in various biological processes, including embryonic development, tissue homeostasis, and cell differentiation.
Overview of Wnt Signaling
Once released by the producing cells, Wnt proteins travel in the extracellular space, to reach their target cells by mechanisms discussed below. In the target cells, Wnt proteins activate signaling by interacting with the extracellular CRD of the cell surface GPCR receptors called Frizzled (Fz). However, in many organisms the complexity of signal activation increases significantly due to the presence of multiple Fz receptors, interacting with a repertoire of Wnt ligands. A combinatorial interaction of different Wnt proteins with Fz receptors leads to the activation of multiple downstream pathways (Figure 1), which are classified into two broad categories based on the involvement of β-catenin: the β-catenin dependent canonical Wnt pathway and the β-catenin independent non-canonical Wnt pathways.
- Canonical Signaling- Wnt-Off: In the absence of the ligand (Wnt), the receptor (Frizzled/Fz) and co-receptor LRP5/6 (Arrow) remain in an inactive state at the plasma membrane. In the cytoplasm, components of the destruction complex (GSK-3β, APC, CK-1, Axin) bind and phosphorylate the nuclear effector β-catenin, followed by its ubiquitination and proteasomal degradation. In the nucleus, transcriptional repressor Groucho (Gro) binds to the co-factor TCF and keeps Wnt target gene expression off.
- Canonical signaling- Wnt-On: Binding of Wnt to the Fz receptor recruits LRP5/6 forming an active receptor complex, which leads to Dishevelled (Dsh)-mediated inactivation of the destruction complex. The consequential accumulation of β-catenin in the cytoplasm enables its nuclear import. The nuclear β-catenin replaces Gro from TCF forming a transcriptional activator complex leading to Wnt target gene expression.
- Non-canonical Wnt/PCP pathway: The binding of Wnt to the Fz receptor leads to the recruitment of Dsh. This complex along with the co-receptors, for example, receptor tyrosine kinase-like orphan receptor (Ror) and tyrosine-protein kinase (Ryk) can activate effector kinases like RhoA/ROCK, which leads to actin polymerization. Wnt proteins can also induce the activation of the c-Jun N-terminal kinase (JNK) pathway through both Fz-Dsh-Ror/Ryk and Vangl-Ror/Ryk protein complexes.
- Non-canonical Wnt/Ca2+ pathway: Fz receptor recruits the co-receptors Ror/Ryk upon binding of Wnt ligands which activates Dsh and G-proteins (α,β) at the membrane forming an active cluster. This results in the activation of phospholipase-C (PLC) leading to the release of intracellular calcium ions. Increased calcium levels further activate different pathways mediated by downstream effectors namely Calcineurin (Cn), Calmodulin dependent protein Kinase II (CAMK II), and Protein Kinase C (PKC).
It's important to note that the Wnt pathway is tightly regulated to ensure proper cellular functions. Dysregulation of Wnt signaling, such as excessive pathway activation or inhibition, can have profound effects on cellular processes and contribute to disease conditions, including cancer and developmental disorders.
Understanding the intracellular signaling events of the Wnt pathway is crucial for developing therapeutic strategies to modulate its activity and restore normal cellular functions in various diseases. Ongoing research continues to unravel the complexity of Wnt signaling and its implications in different biological contexts.
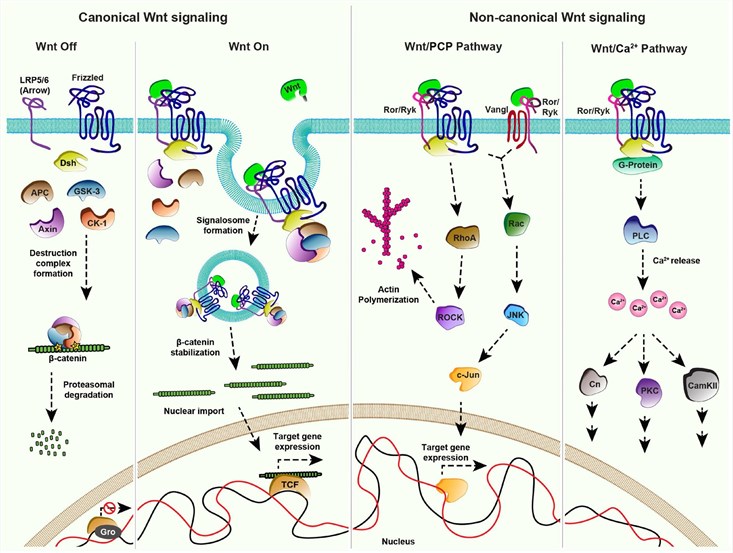 Fig.1 Wnt signaling pathways. (Mehta S, et al., 2021)
Fig.1 Wnt signaling pathways. (Mehta S, et al., 2021)
Key Molecules in Wnt Intracellular Signaling
Here are some key molecules involved in Wnt intracellular signaling, such as Wnt ligands, Frizzled receptors, Co-receptors, Disheveled (DVL), β-catenin, Axin, TCF/LEF transcription factors, GSK3β. The intricate interactions and regulatory mechanisms involving these molecules contribute to the complexity and versatility of the Wnt signaling pathway. Further research continues to uncover additional components and mechanisms that modulate Wnt signaling and its implications in various biological processes.
Functions of Wnt Intracellular Signaling
Wnt intracellular signaling plays crucial roles in various cellular processes and developmental events. Here are some key functions of Wnt intracellular signaling:
- Cell Fate Specification and Differentiation: Wnt signaling is involved in determining cell fate during embryonic development and tissue homeostasis. It regulates the differentiation of stem cells into specific cell types by activating or repressing the expression of lineage-specific genes. Wnt signaling can direct cells towards different developmental pathways, influencing organogenesis and tissue formation.
- Cell Proliferation and Growth: Wnt signaling is closely linked to cell proliferation and growth. In many tissues, Wnt signaling promotes cell division and tissue expansion. It regulates the cell cycle by influencing the expression of genes involved in cell cycle control, such as cyclins and cyclin-dependent kinases (CDKs). Dysregulation of Wnt signaling can lead to uncontrolled cell proliferation, contributing to tumor formation and cancer progression.
- Tissue Regeneration and Repair: Wnt signaling plays a critical role in tissue regeneration and repair processes. Upon tissue damage, Wnt signaling is activated to initiate the proliferation and migration of nearby cells, promoting tissue repair. It is involved in the activation of resident stem cells and the recruitment of progenitor cells to the site of injury, facilitating tissue regeneration.
- Synaptic Plasticity and Neurodevelopment: Wnt signaling is essential for proper neuronal development and synaptic plasticity—the ability of synapses to modify their strength and connectivity in response to activity and experience. Wnt signaling regulates axon guidance, dendrite formation, and synapse development in the nervous system. It also influences the functional maturation and maintenance of synapses.
- Planar Cell Polarity: In addition to the canonical Wnt signaling pathway, non-canonical Wnt signaling pathways, such as the planar cell polarity (PCP) pathway, regulate the orientation and polarity of cells within a tissue plane. PCP signaling is involved in processes such as cell migration, tissue morphogenesis, and the alignment of hairs or cilia on cells. It coordinates cellular behaviors to achieve proper tissue organization.
- Stem Cell Maintenance and Self-Renewal: Wnt signaling plays a critical role in the self-renewal and maintenance of various stem cell populations. It helps to maintain the undifferentiated state of stem cells and regulates their proliferation and differentiation. Proper regulation of Wnt signaling is crucial for the balance between self-renewal and differentiation of stem cells.
- Disease Pathogenesis: Dysregulation of Wnt signaling has been implicated in various diseases, including cancer, neurodegenerative disorders, and developmental abnormalities. Aberrant activation or inhibition of the pathway can contribute to pathological conditions. Understanding and modulating Wnt signaling holds potential for therapeutic interventions in these diseases.
It's important to note that the functions of Wnt intracellular signaling can vary depending on the cellular context and the specific downstream effectors activated by the pathway. The intricate regulation and cross-talk of Wnt signaling with other signaling pathways contribute to its diverse functions in development, homeostasis, and disease.
If you have any questions, requirements, or cooperation intentions, please feel free to contact us. We very much look forward to working with you and helping you achieve research and commercial success.
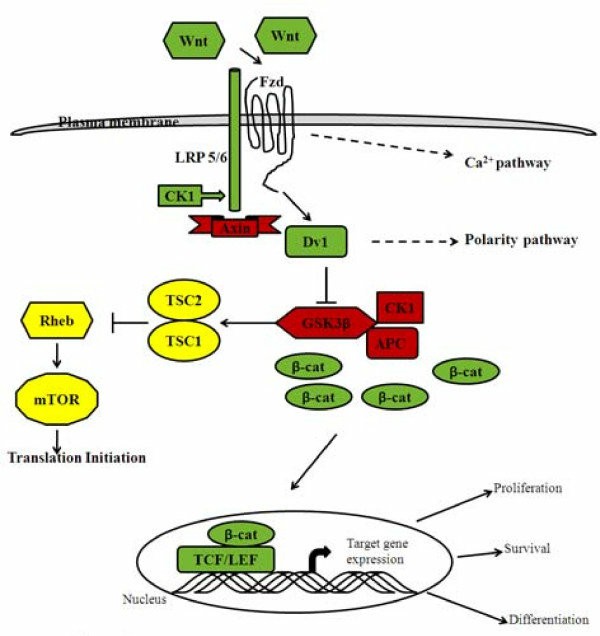 Fig.2 Schematic representation of the Wnt signaling pathway in cancer cells. (Majid S, et al., 2012)
Fig.2 Schematic representation of the Wnt signaling pathway in cancer cells. (Majid S, et al., 2012)
In the presence of active Wnt, β-catenin accumulates in the cytoplasm, then localizes to the nucleus, and activates transcription together with TCF/LEF transcription factors. Negative regulators are depicted in red and positive regulators in green. Activation of the mTOR pathway is also directly regulated by Wnt-dependent downregulation of GSK3 kinase activity which is depicted in yellow.
References:
- Mehta S, Hingole S, Chaudhary V. The Emerging Mechanisms of Wnt Secretion and Signaling in Development. Front Cell Dev Biol. 2021;9:714746. Published 2021 Aug 16. doi:10.3389/fcell.2021.7147467
- Majid S, Saini S, Dahiya R. Wnt signaling pathways in urological cancers: past decades and still growing. Mol Cancer. 2012;11:7. Published 2012 Feb 10. doi:10.1186/1476-4598-11-7

